Merge and Mark Duplicates¶
mergeAndMarkBams · 1 contributor · 3 versions
No documentation was provided: contribute one
Quickstart¶
from janis_bioinformatics.tools.common.mergeandmark.mergeandmark_4_1_3 import MergeAndMarkBams_4_1_3 wf = WorkflowBuilder("myworkflow") wf.step( "mergeandmarkbams_step", MergeAndMarkBams_4_1_3( bams=None, ) ) wf.output("out", source=mergeandmarkbams_step.out)
OR
- Install Janis
- Ensure Janis is configured to work with Docker or Singularity.
- Ensure all reference files are available:
Note
More information about these inputs are available below.
- Generate user input files for mergeAndMarkBams:
# user inputs
janis inputs mergeAndMarkBams > inputs.yaml
inputs.yaml
bams:
- bams_0.bam
- bams_1.bam
- Run mergeAndMarkBams with:
janis run [...run options] \
--inputs inputs.yaml \
mergeAndMarkBams
Information¶
URL: No URL to the documentation was provided
| ID: | mergeAndMarkBams |
|---|---|
| URL: | No URL to the documentation was provided |
| Versions: | 4.0.12, 4.1.2, 4.1.3 |
| Authors: | Michael Franklin |
| Citations: | |
| Created: | 2019-02-19 |
| Updated: | 2020-11-06 |
Outputs¶
| name | type | documentation |
|---|---|---|
| out | IndexedBam |
Workflow¶
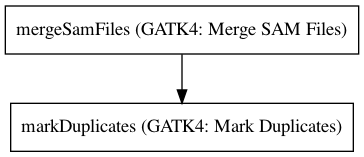
Embedded Tools¶
| GATK4: Merge SAM Files | Gatk4MergeSamFiles/4.1.3.0 |
| GATK4: Mark Duplicates | Gatk4MarkDuplicates/4.1.3.0 |
Additional configuration (inputs)¶
| name | type | documentation |
|---|---|---|
| bams | Array<IndexedBam> | |
| createIndex | Optional<Boolean> | |
| maxRecordsInRam | Optional<Integer> | |
| sampleName | Optional<String> | |
| mergeSamFiles_useThreading | Optional<Boolean> | Option to create a background thread to encode, compress and write to disk the output file. The threaded version uses about 20% more CPU and decreases runtime by ~20% when writing out a compressed BAM file. |
| mergeSamFiles_validationStringency | Optional<String> | Validation stringency for all SAM files read by this program. Setting stringency to SILENT can improve performance when processing a BAM file in which variable-length data (read, qualities, tags) do not otherwise need to be decoded.The –VALIDATION_STRINGENCY argument is an enumerated type (ValidationStringency), which can have one of the following values: [STRICT, LENIENT, SILENT] |
Workflow Description Language¶
version development
import "tools/Gatk4MergeSamFiles_4_1_3_0.wdl" as G
import "tools/Gatk4MarkDuplicates_4_1_3_0.wdl" as G2
workflow mergeAndMarkBams {
input {
Array[File] bams
Array[File] bams_bai
Boolean? createIndex = true
Int? maxRecordsInRam = 5000000
String? sampleName
Boolean? mergeSamFiles_useThreading = true
String? mergeSamFiles_validationStringency = "SILENT"
}
call G.Gatk4MergeSamFiles as mergeSamFiles {
input:
bams=bams,
bams_bai=bams_bai,
sampleName=sampleName,
useThreading=select_first([mergeSamFiles_useThreading, true]),
createIndex=select_first([createIndex, true]),
maxRecordsInRam=select_first([maxRecordsInRam, 5000000]),
validationStringency=select_first([mergeSamFiles_validationStringency, "SILENT"])
}
call G2.Gatk4MarkDuplicates as markDuplicates {
input:
bam=[mergeSamFiles.out],
outputPrefix=sampleName,
createIndex=select_first([createIndex, true]),
maxRecordsInRam=select_first([maxRecordsInRam, 5000000])
}
output {
File out = markDuplicates.out
File out_bai = markDuplicates.out_bai
}
}
Common Workflow Language¶
#!/usr/bin/env cwl-runner
class: Workflow
cwlVersion: v1.2
label: Merge and Mark Duplicates
doc: ''
requirements:
- class: InlineJavascriptRequirement
- class: StepInputExpressionRequirement
- class: MultipleInputFeatureRequirement
inputs:
- id: bams
type:
type: array
items: File
secondaryFiles:
- pattern: .bai
- id: createIndex
type: boolean
default: true
- id: maxRecordsInRam
type: int
default: 5000000
- id: sampleName
type:
- string
- 'null'
- id: mergeSamFiles_useThreading
doc: |-
Option to create a background thread to encode, compress and write to disk the output file. The threaded version uses about 20% more CPU and decreases runtime by ~20% when writing out a compressed BAM file.
type: boolean
default: true
- id: mergeSamFiles_validationStringency
doc: |-
Validation stringency for all SAM files read by this program. Setting stringency to SILENT can improve performance when processing a BAM file in which variable-length data (read, qualities, tags) do not otherwise need to be decoded.The --VALIDATION_STRINGENCY argument is an enumerated type (ValidationStringency), which can have one of the following values: [STRICT, LENIENT, SILENT]
type: string
default: SILENT
outputs:
- id: out
type: File
secondaryFiles:
- pattern: .bai
outputSource: markDuplicates/out
steps:
- id: mergeSamFiles
label: 'GATK4: Merge SAM Files'
in:
- id: bams
source: bams
- id: sampleName
source: sampleName
- id: useThreading
source: mergeSamFiles_useThreading
- id: createIndex
source: createIndex
- id: maxRecordsInRam
source: maxRecordsInRam
- id: validationStringency
source: mergeSamFiles_validationStringency
run: tools/Gatk4MergeSamFiles_4_1_3_0.cwl
out:
- id: out
- id: markDuplicates
label: 'GATK4: Mark Duplicates'
in:
- id: bam
source:
- mergeSamFiles/out
linkMerge: merge_nested
- id: outputPrefix
source: sampleName
- id: createIndex
source: createIndex
- id: maxRecordsInRam
source: maxRecordsInRam
run: tools/Gatk4MarkDuplicates_4_1_3_0.cwl
out:
- id: out
- id: metrics
id: mergeAndMarkBams