Welcome to Janis!¶


Note
This project is work-in-progress and is provided as-is without warranty of any kind. There may be breaking changes committed to this repository without notice.
It was developed as part of the Portable Pipelines Project, a collaboration between:
- Melbourne Bioinformatics (University of Melbourne)
- Peter MacCallum Cancer Centre
- Walter and Eliza Hall Institute of Medical Research (WEHI).
Janis has two major functionalities:
- Translation between workflow languages, and
- Modelling tools and workflows using a Python API.
Janis requires a Python installation > 3.10, and can be installed through PIP project page by running:
pip install janis-pipelines
— Translation —¶
Overview¶
Janis can read-in from CWL, WDL, and Galaxy, and can write-out to CWL, WDL, and Nextflow.
To facilitate translation, Janis includes an internal API to model tools and workflows in a way that encompasses the design features of each supported language. When performing a translation, Janis will ingest the source files into the internal API, then will translate out to the target language.
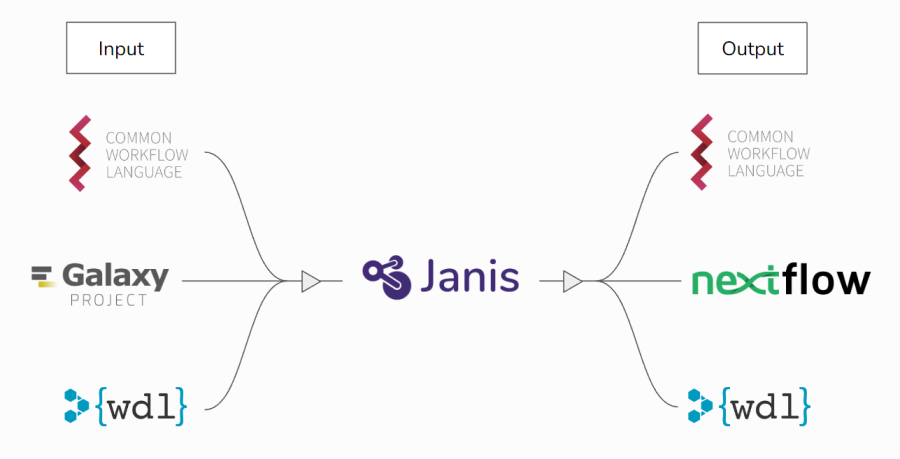
Janis produces translations as close to the original as possible, but in some situations it may be necessary to make manual adjustments before the output is runnable. This is due to language-specific features of supported languages, and their inclusion of freeform Javascript (CWL), Java / Groovy (Nextflow), and Python / Cheetah (Galaxy).
Quickstart¶
To try out Janis on a sample tool and workflow, follow the steps below.
Install Janis
python -m venv venv
source venv/bin/activate
pip install janis-pipelines
Obtain Sample Files
git clone https://github.com/GraceAHall/sample_translation_files
Sample Tool Translation: CWL to Nextflow
janis translate --from cwl --to nextflow sample_translation_files/cwl_tool/samtools_flagstat.cwl
Sample Workflow Translation: CWL to Nextflow
janis translate --from cwl --to nextflow sample_translation_files/cwl_workflow/align_sort_markdup.cwl
Tutorials¶
Janis currently has full tutorials for translations of CWL -> Nextflow, and Galaxy -> Nextflow.
CWL to Nextflow
Galaxy to Nextflow
— Python API —¶
Overview¶
The internal API used by Janis can be directly used to model bioinformatics tools and workflows using Python. To date, dozens of bioinformatics pipelines are written purely using the Janis API.
To execute these pipelines researchers use Janis’ translate ability to produce CWL / WDL which is then run on the target platform used by their institution.
These pipelines are available in the janis-pipelines repository.
Example Workflow¶
# write the workflow to `helloworld.py`
cat <<EOT >> helloworld.py
import janis as j
from janis_unix.tools import Echo
w = j.WorkflowBuilder("hello_world")
w.input("input_to_print", j.String)
w.step("echo", Echo(inp=w.input_to_print))
w.output("echo_out", source=w.echo.out)
EOT
# Translate workflow to WDL
janis translate helloworld.py wdl
# Run the workflow
janis run -o helloworld-tutorial helloworld.py --input_to_print "Hello, World!"
# See your output
cat helloworld-tutorial/echo_out
# Hello, World!
Tutorials¶
Workshops¶
In addition, there are fully self-guided workshops that more broadly go through the functionality of Janis:
Examples¶
Sometimes it’s easier to learn by examples, here are a few hand picked examples:
Toolbox¶
There are two toolboxes currently available on Janis:
- Unix <https://github.com/PMCC-BioinformaticsCore/janis-unix>`__(`list of tools)
- Bioinformatics <https://github.com/PMCC-BioinformaticsCore/janis-bioinformatics>`__(`list of tools)
— Additional —¶
References¶
Through conference or talks, this project has been referenced by the following titles:
- Walter and Eliza Hall Institute Talk (WEHI) 2019: Portable Pipelines Project: Developing reproducible bioinformatics pipelines with standardised workflow languages
- Bioinformatics Open Source Conference (BOSC) 2019: Janis: an open source tool to machine generate type-safe CWL and WDL workflows
- Victorian Cancer Bioinformatics Symposium (VCBS) 2019: Developing portable variant calling pipelines with Janis
- GIW / ABACBS 2019: Janis: A Python framework for Portable Pipelines
- Australian BioCommons, December 2019: Portable pipelines: build once and run everywhere with Janis
Support¶
To get help with Janis, please ask a question on Gitter or raise an issue on GitHub.
If you find an issue with the tool definitions, please see the relevant issue page:
This project is work-in-progress and is still in developments. Although we welcome contributions, due to the immature state of this project we recommend raising issues through the Github issues page for Pipeline related issues.
Information about the project structure and more on contributing can be found within the documentation.